Deck: If the wall breathes in Part 2, the next step is to give its chemistry a memory and a reason to persist—by packaging it into protocells, copying with variation, and letting selection do the rest.
First, the interface must warm itself (autocatalysis + exotherm). A spark is not a creature. It needs a body—and a way to remember.
TL;DR
- We will feed vesicles with the interface‑heartbeat effluent and measure whether cargo‑rich protocells grow/divide faster than empty ones—and keep the edge after reseeding. (Until the effluent is available, we use a matched synthetic mix.)
- In parallel, we run templated copying under cycles (wet–dry or temperature) using planet‑plausible activation (AcP/PPi; metals), record error spectra, and couple it to compartments.
- Pass = (1) multi‑cycle advantage that persists after reseed, and (2) replication with heritable variation. Together with a successful heartbeat dataset, that would cross the Darwin threshold with no enzymes or one‑off miracles.
Controls include UMI barcodes, no-carry-over sentinels, and size-matched competitions to rule out artefacts.
Where Part 2 leaves off (30‑second recap)
Part 2 defines a single pass/fail experiment—the Interface‑Heartbeat—at a porous FeS core / Fe‑phosphate rim wall. A passing run would show (i) a sigmoidal CO₂→C₂–C₄ curve and (ii) a matching exotherm from the same interface, with small ΔpH/ΔE bumps and AcP/PPi formation. When that dataset lands, the spark becomes mechanistic.
Today’s question (conditional): if the spark lights, can it inherit?
The wager
If chemistry can copy with variation inside a container, then selection is inevitable. We’ll ask three things of a prebiotic‑plausible system:
- Package: Do protocells that inherit useful cargo outgrow and outlast empty ones?
- Copy: Can short polymers template their own extension across ≥3 cycles, with a measurable error profile?
- Select: Does the advantage persist after reseeding into fresh media?
If yes to all three, we have protolife—the minimal engine of evolution.
Scene 1 — Bottling the heartbeat (feeds)
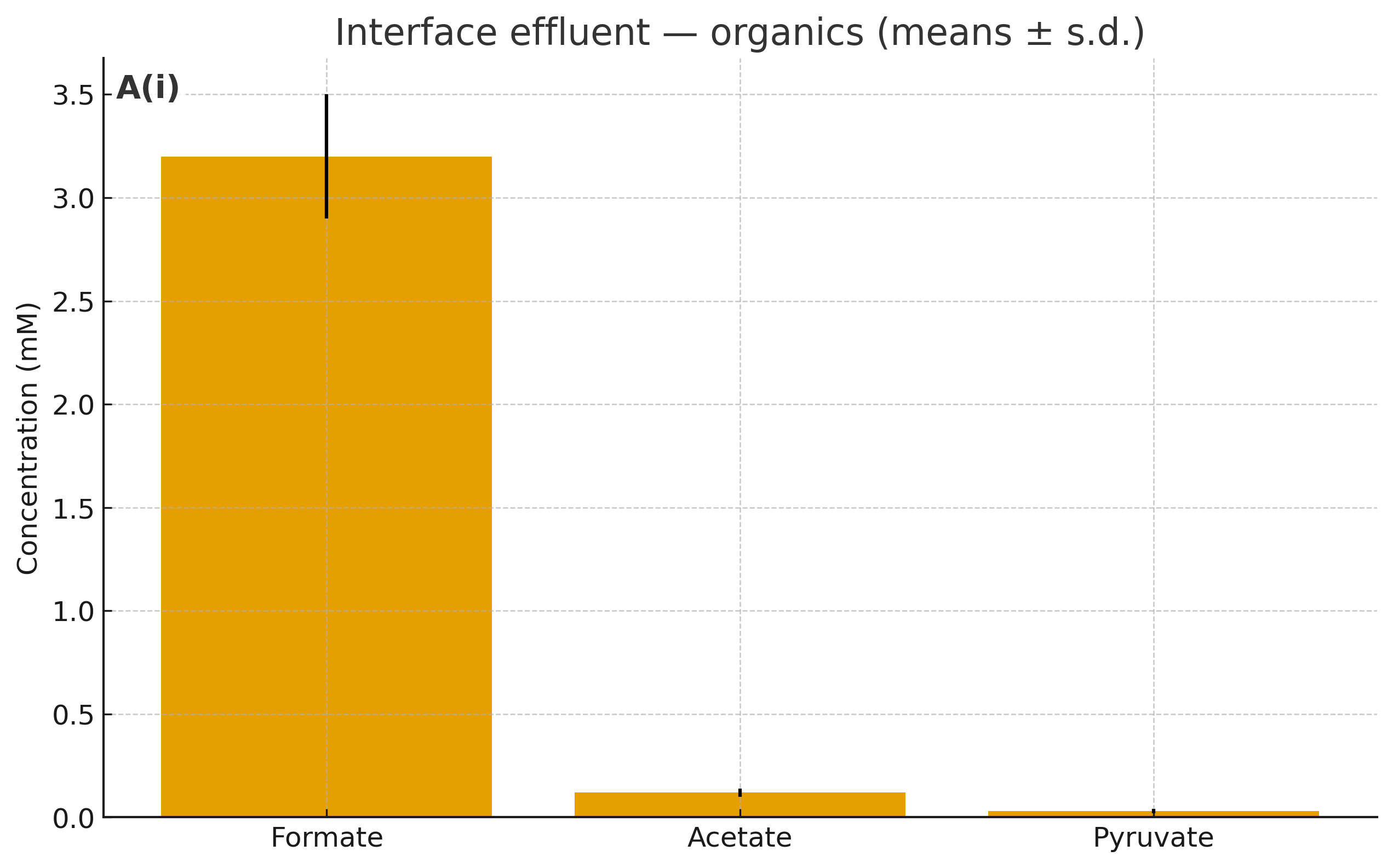
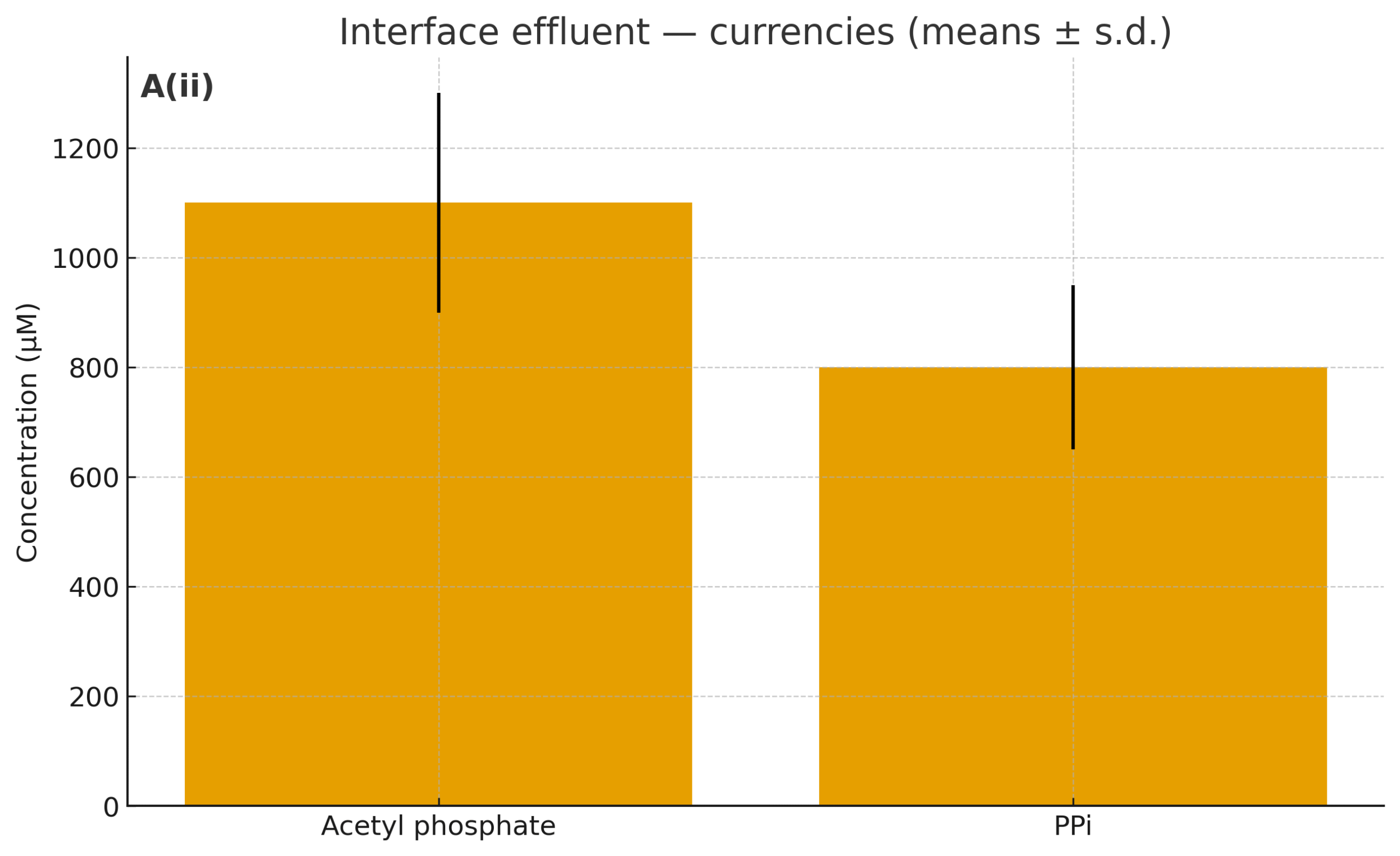
We will treat the interface reactor as a source of simple carbon and currencies, then run a split (while the interface run is pending, we start with synthetic mixes matched to the target composition):
-
Effluent fractions: 1×, 0.3×, 0.1×. Each fraction is filtered/sterile‑handled.
-
What’s inside (typical ranges): formate (mM), acetate (µM–mM), ±pyruvate (µM), AcP/PPi (sub‑µM–µM), K⁺/Mg²⁺/PO₄³⁻ salts, trace Fe²⁺/Ni²⁺.
-
Controls: matched synthetic mixes reproducing the measured composition; and “blank” effluent from control walls.
Scene 2 — Membranes that can live here (containers)
We use fatty-acid:glycerol-monoester (FA:GME) membranes (~2:1) at pH 8–9. With Mg²⁺ chelated by citrate (≈1:1–1:2), these vesicles grow and divide and tolerate the K⁺/Mg²⁺ windows we target; salts are tuned to the Interface-Heartbeat regime. All copying runs use Mg²⁺–citrate.
Setup
- Load vesicles by gentle hydration with each effluent fraction (and synthetic‑mix controls).
- Monitor size (DLS/EM), division (time‑lapse), and cargo retention (fluorescent standards where relevant).
- Cycles: chemostat‑style dilution or wet–dry rehydration appropriate to the salt window.
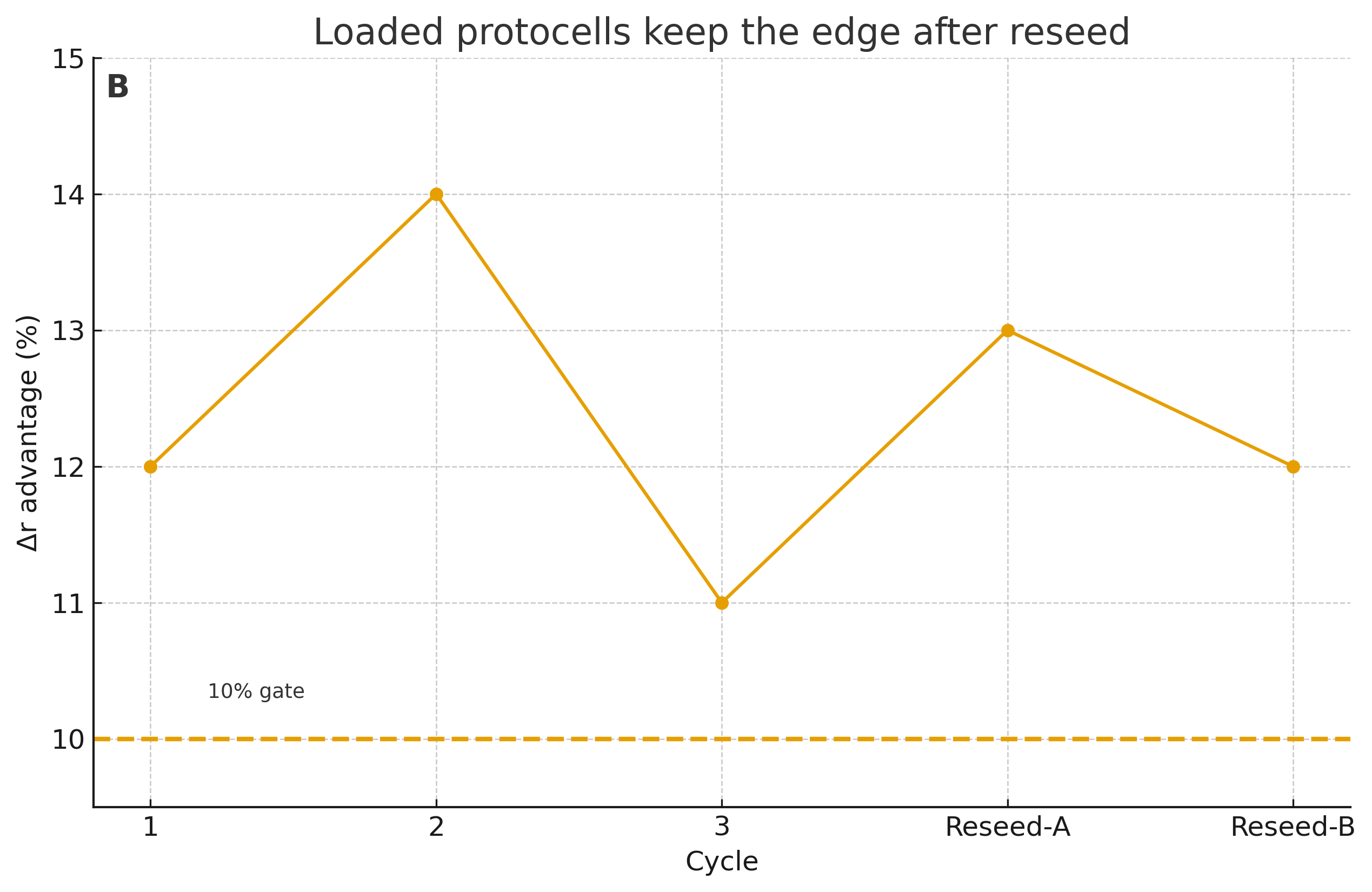
Pass (advantage): cargo-rich protocells show Δr ≥ 10% over ≥3 cycles vs empty controls, in size-matched competitions, and retain the edge after reseeding.
Failure diagnostics
- Advantage only in the first cycle → check osmotic transients; tune membrane recipe (FA:GME ratio).
- Growth but no division → extend cycles or increase shear; add amphiphile feed.
Scene 3 — Teaching chemistry to remember (copying under cycles)
We run two complementary lanes using the same salts and temperatures:
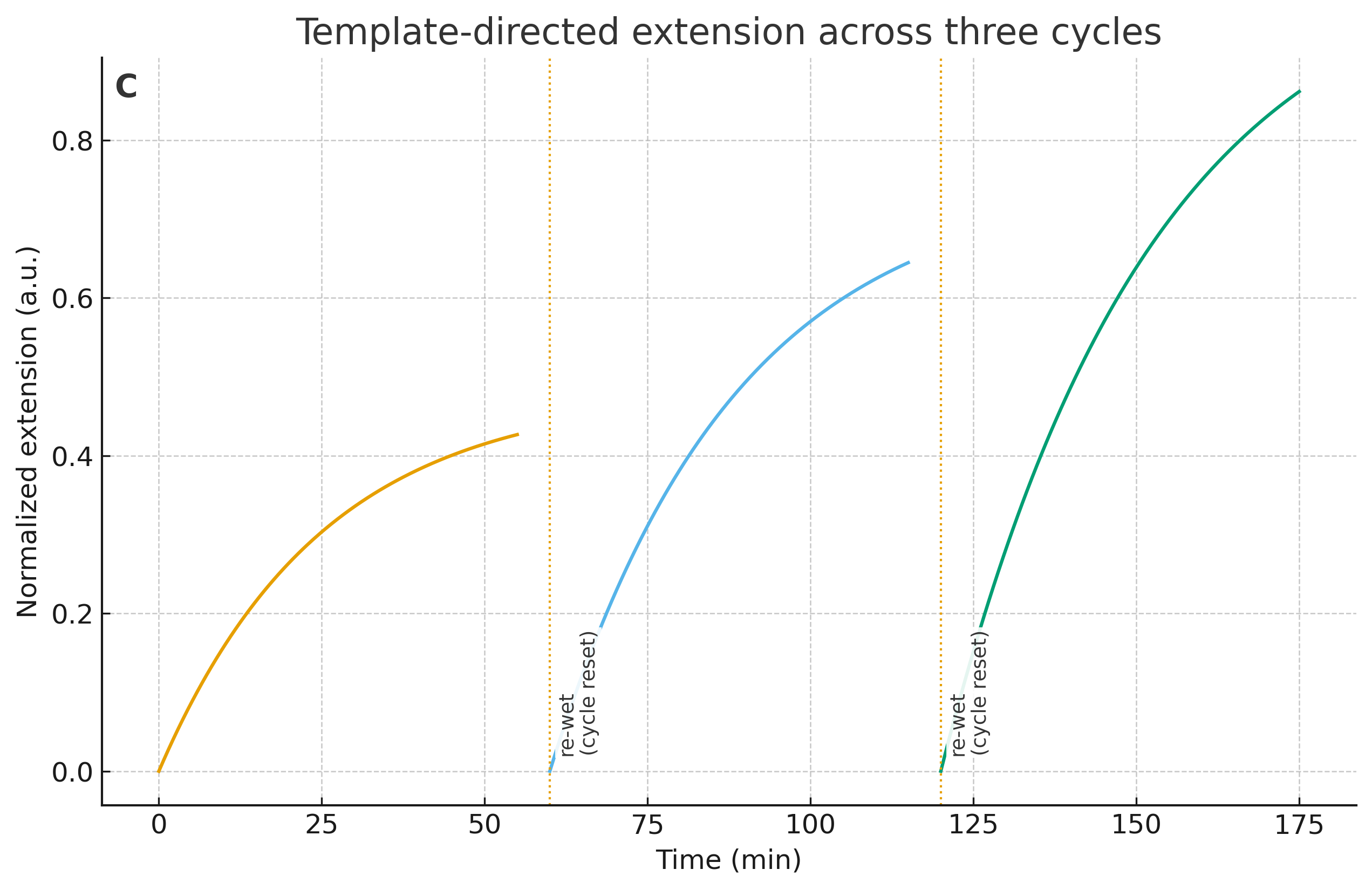
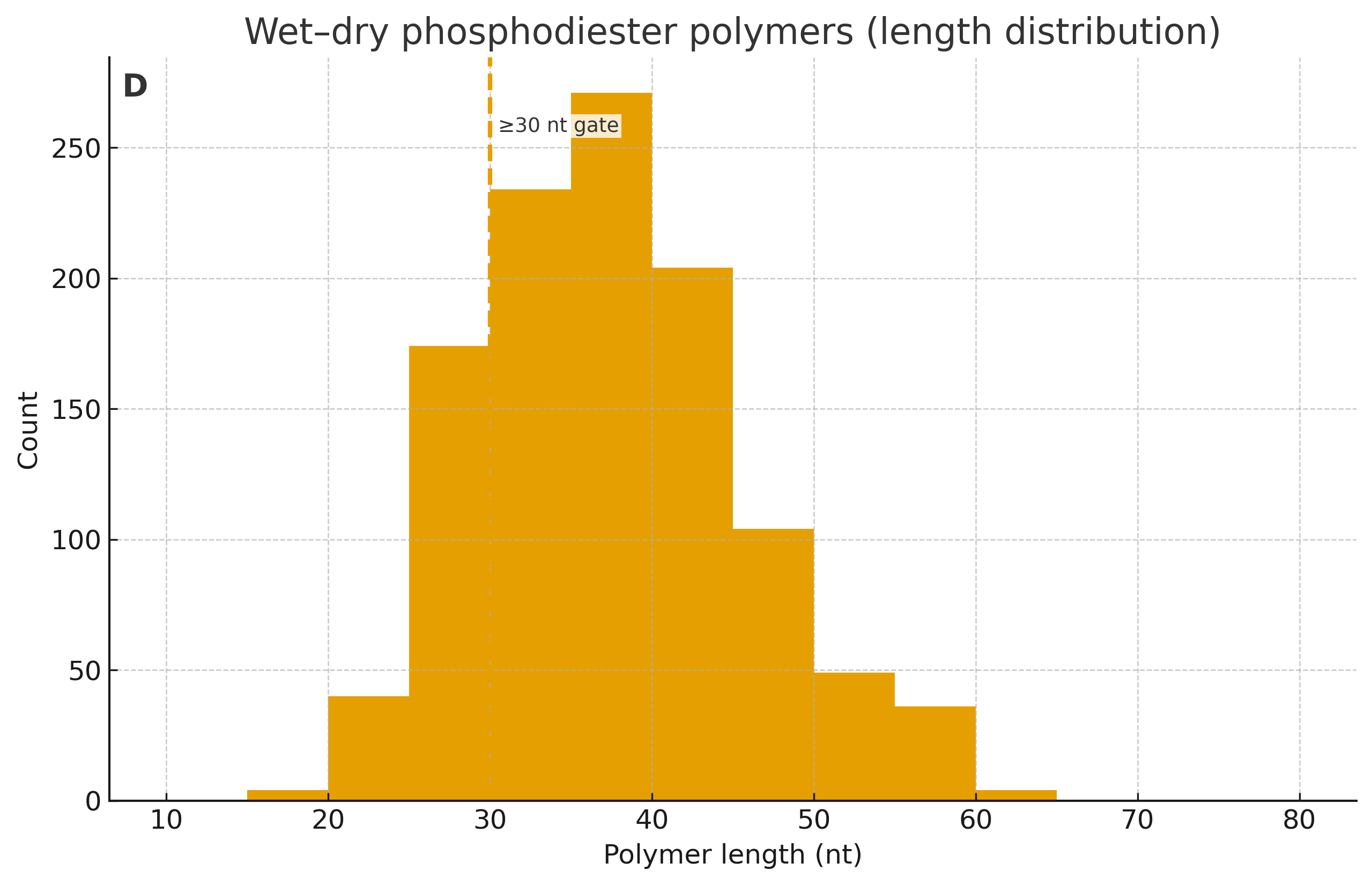
- Wet–dry stitching (polymerization): drive true phosphodiester linkages to tens of nt on dry-down; re-wet via limited rehydration (humidity/deliquescent salts, not bulk water); repeat ≥3 cycles. Quantify length distributions by PAGE/CE/LC-MS.
- Template‑directed extension/ligation: short templates (e.g., 10–20 nt) extended or ligated under temperature or wet–dry cycles.
Activation hierarchy (prebiotic‑lean → method control):
- Primary: AcP/PPi‑linked phosphorylation/ligation with Fe²⁺/Zn²⁺ assistance.
- Control: 2‑aminoimidazole (2‑AI) protocols run as method controls only, not headline.
Targets
- ≥30-nt phosphodiester oligomers (often mixed 3′–5′/2′–5′) confirmed by nuclease/enzymatic digestion signatures.
- ≥3 cycles with carry‑over and error spectra (deep sequencing) to show heritability of variation.
Use UMI barcodes to deduplicate and count unique copied/ligated molecules.
Coupling to compartments
- Run copying inside loaded vs empty vesicles; track whether cargo improves either copying yield or growth. Selection should favor the better copier/grower. Feed activated monomers externally; Mg²⁺–citrate increases permeability to short oligomers while protecting membranes.
Failure diagnostics
- Only 2‑AI works → raise AcP/PPi supply from heartbeat reactor; adjust Mg²⁺ and pH; check for phosphate buffering that quenches activation.
- Copying without heritable errors (noise) → increase cycle number and sequence complexity; ensure carry‑over is real (no full reset).
Scene 4 — The first tournament (selection)
We pit loaded vs empty protocells in round‑robin competitions:
- Schedule: three growth-and-division cycles, then reseed (10% inoculum) into fresh media for two more cycles.
- Metrics: size distributions, division counts, internal cargo retention, Δr per cycle, and sequence data for heritable variation.
- Decision rule (pass):
- Δr ≥ 10% in loaded vs empty in size-matched competitions across ≥3 cycles;
- advantage persists after reseed; and
- replication with heritable variation is is confirmed by UMI-aware sequence analysis.
Methods at a glance (for readers who want to run it)
Membranes
FA:GME ≈2:1; pH 8–9; K⁺/Mg²⁺ regime; optional short peptides from wet–dry lane as co‑surfactants.
Copying
Wet–dry: 60–90 °C dry; 20–40 min intervals; ≥3 cycles; limited rehydration.
Template extension/ligation: gentle temperature cycles; AcP/PPi (prebiotic) with metals where indicated; 2-AI is method control only; use 2′,3′-cNMPs where available.
Analytics
DLS/EM for membranes; PAGE/CE/LC‑MS for polymers; deep sequencing for errors; fluorimetry for encapsulation; simple OD/absorbance or particle counting for growth.
Controls
Empty vesicles; synthetic‑mix cargo; activation‑chemistry blanks; “copying outside vesicles” controls; no‑carry‑over controls; and a 2‑AI method control clearly labeled as such.
Stats & preregistration
Pre‑register Δr threshold (≥10%), cycle count (≥3 + reseed), replication length (≥30 nt), and an analysis plan for error spectra. Share code and CSV schemas.
Pass/fail table (short form)
| Gate | Pass | Fail |
|---|---|---|
| Packaging advantage | Loaded protocells show Δr ≥ 10% for ≥3 cycles and keep the edge after reseed | Advantage vanishes after reseed; advantage only with exotic activators |
| Copy with variation | ≥30‑nt polymers and templated extension/ligation with heritable error spectra across cycles | Only short oligomers; copying resets each cycle; no heritable pattern |
| Coupled selection | Copying inside loaded > inside empty protocells | No difference between compartments |
Together with a successful heartbeat proof (S‑curve + exotherm + currencies), this would cross the Darwin threshold.
Fail‑fast rituals (when something’s off)
-
Growth without division: increase shear or amphiphile feed; adjust FA:GME ratio.
-
Division without advantage: raise currency flux from the heartbeat source or concentrate effluent.
-
Copying only with 2‑AI: verify AcP/PPi availability; reduce phosphate buffering; tune Mg²⁺/pH.
-
Advantage but no reseed persistence: the edge is environmental; re‑optimize loading so the capability is inherited, not borrowed.
Safety & scope
No genes, no pathogens, no chassis. Prebiotic salts and small molecules only. Waste neutralized/alkaline. This is a chemistry experiment about the minimum engine of evolution.
Why this matters
Life didn’t begin with a blueprint. It began with interfaces that did work, containers that held onto useful stuff, and chemistry that could almost remember—until it finally could. If we can show copying with variation inside a protocell that wins competitions, and if the interface heartbeat is demonstrated as planned, selection will do what selection does.
