Deck (standfirst): If the wall “heartbeat” (Part 2) passes and our protocell copying (Part 3) clears the bar, the next step is to feed that system deoxynucleosides made by daylight chemistry and watch whether deoxy content rises cycle by cycle. We stay enzyme-free: sulfite/H₂S + UV-B makes the feed; wet–dry (or thermal) cycles do the copying; protocell selection keeps score.
TL;DR
-
Make the deoxy feed with UV-B + sulfite from thioanhydro precursors (pH 8–9).
-
Copy under cycles with mixed rN+dN activated monomers; quantify deoxy fraction per round.
-
Selection check: loaded protocells keep a ≥10% growth edge after reseeds.
-
Pass if: (i) UV-B-only bandpass, (ii) LC-MS shows dA/dI in the tens-µM range, (iii) deoxy fraction slope > 0 (95% CI), (iv) Δr ≥ 10% persists after reseed and (v) cleanup thresholds are met (sulfite ≤ 0.5–1 mM; Fe ≤ 10 µM)
Why this matters (one bridge, not a miracle)
Prebiotic photoredox under UV-B with sulfite or H₂S can selectively reduce ribo precursors to deoxy purines (e.g., dA, dI) at mild pH. That gives a geochemically plausible DNA-leaning feed. If those deoxynucleosides join non-enzymatic copying and their fraction grows each cycle, we’ve shown a directional path from RNA-first chemistry toward mixed RNA/DNA—no enzymes, no lab magic.
Experiment outline (what we’ll actually do)
A) Make the deoxy feed (UV-B photoredox, sulfite)
-
Setup. 10 mM carbonate (preferred) or borate buffer, pH 8.3–8.8; 5 mM thioanhydro purine precursor; 50 mM NaHSO₃ (or Na₂SO₃).
-
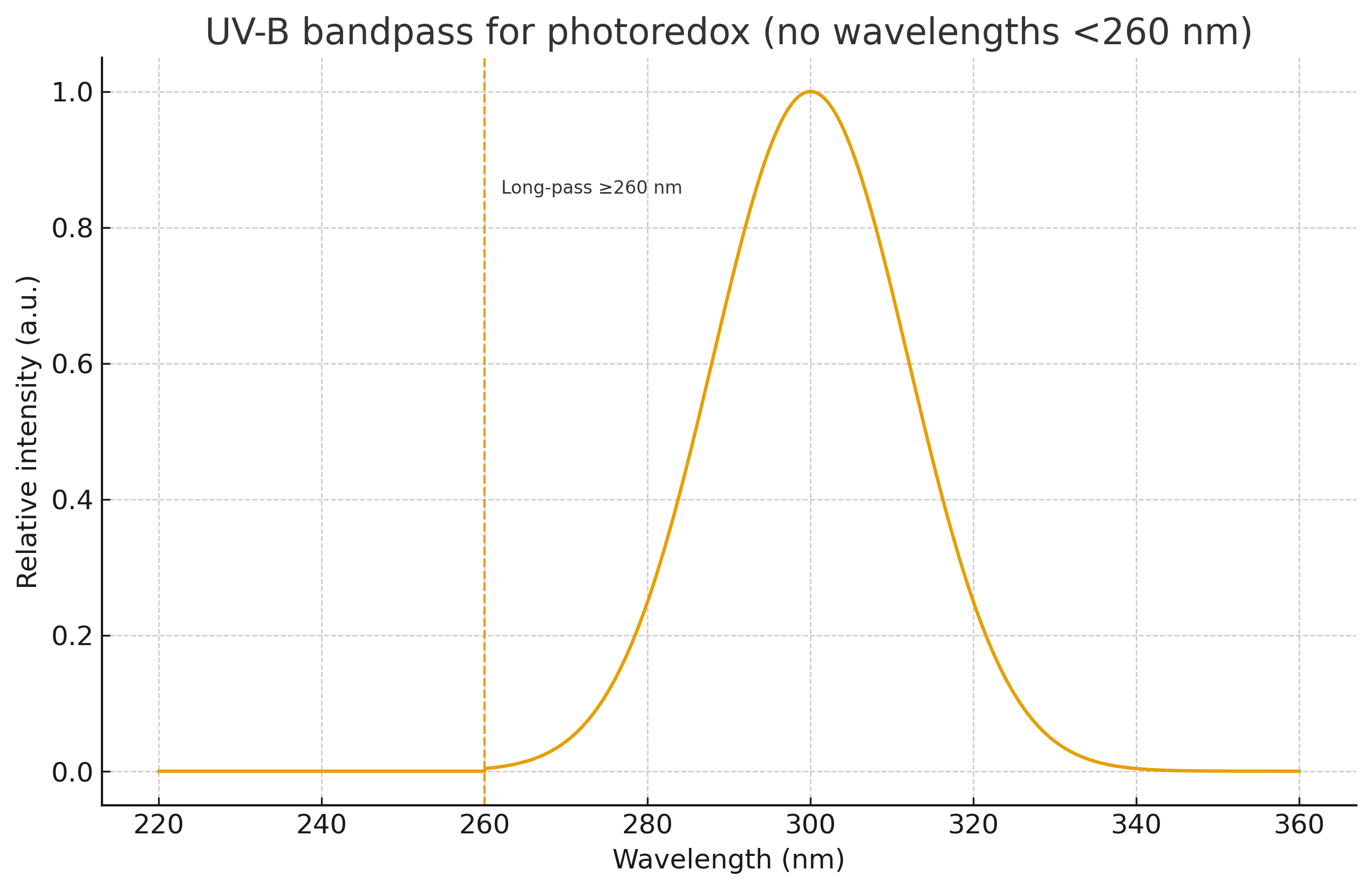
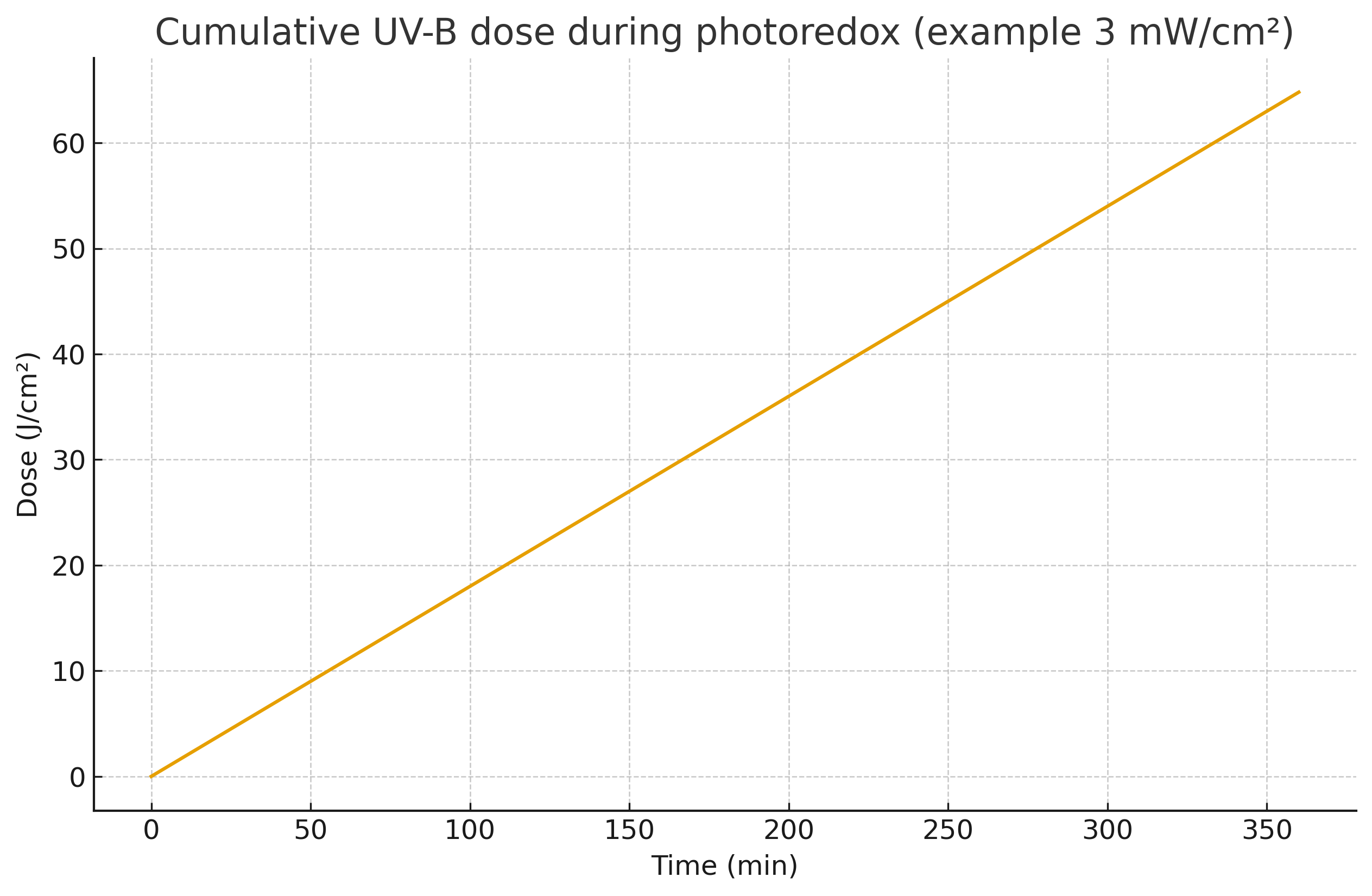
Illumination. UV-B 280–320 nm, long-pass ≥260 nm, 2–5 mW cm⁻², 25–35 °C, 4–8 h.
-
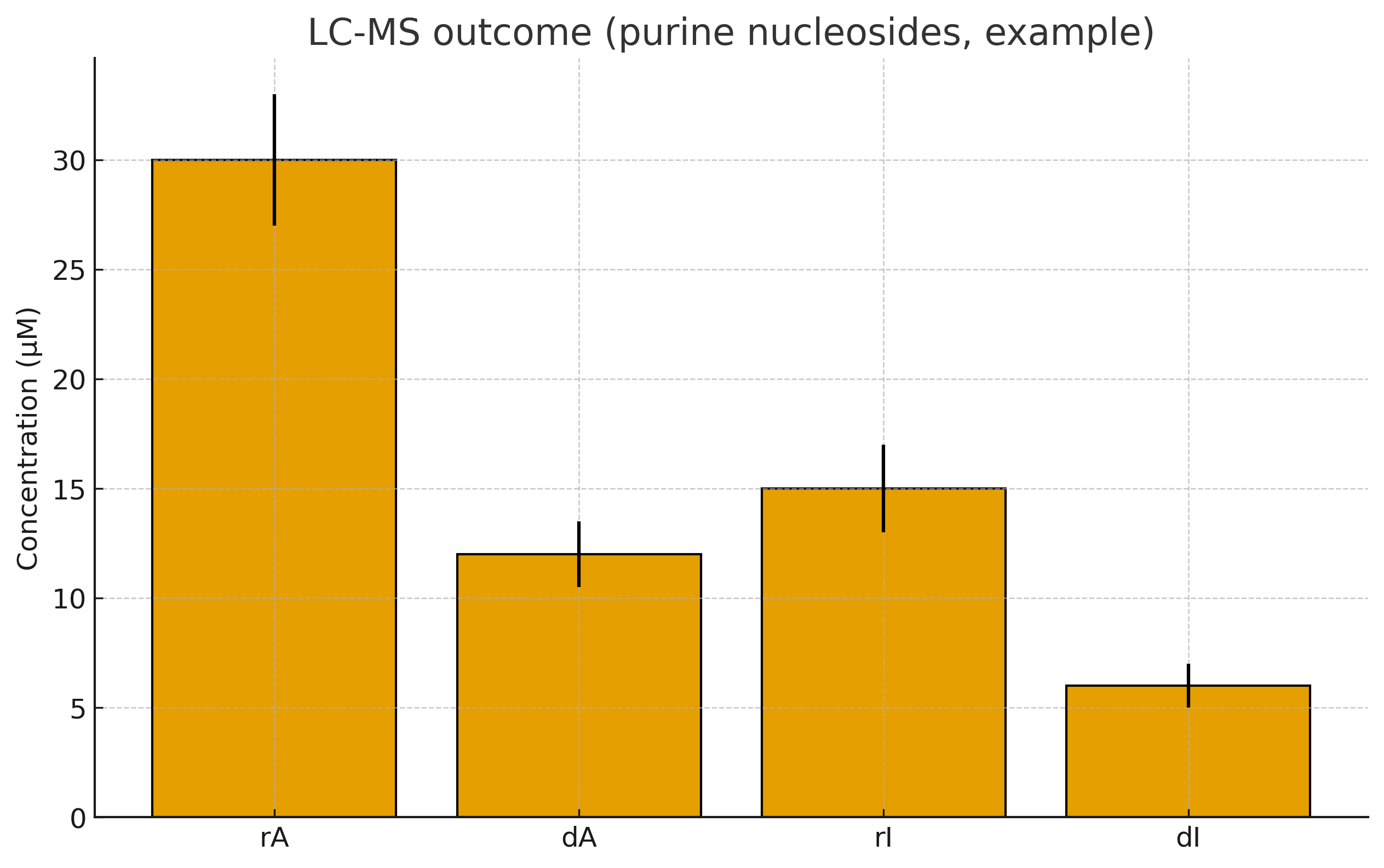
Readout. LC-MS quant for rA/dA and rI/dI; report absolute µM and dN:rN ratios.
A-pass gate (feed):
-
Bandpass verified UV-B-only; sub-260 nm irradiance ≤ 1% of the UV-B band (figure A1a).
-
Cumulative dose 20–70 J cm⁻² (A1b).
-
LC-MS detects dA or dI ≥5–10 µM each (example A2); total dN:rN ≥ 0.3.
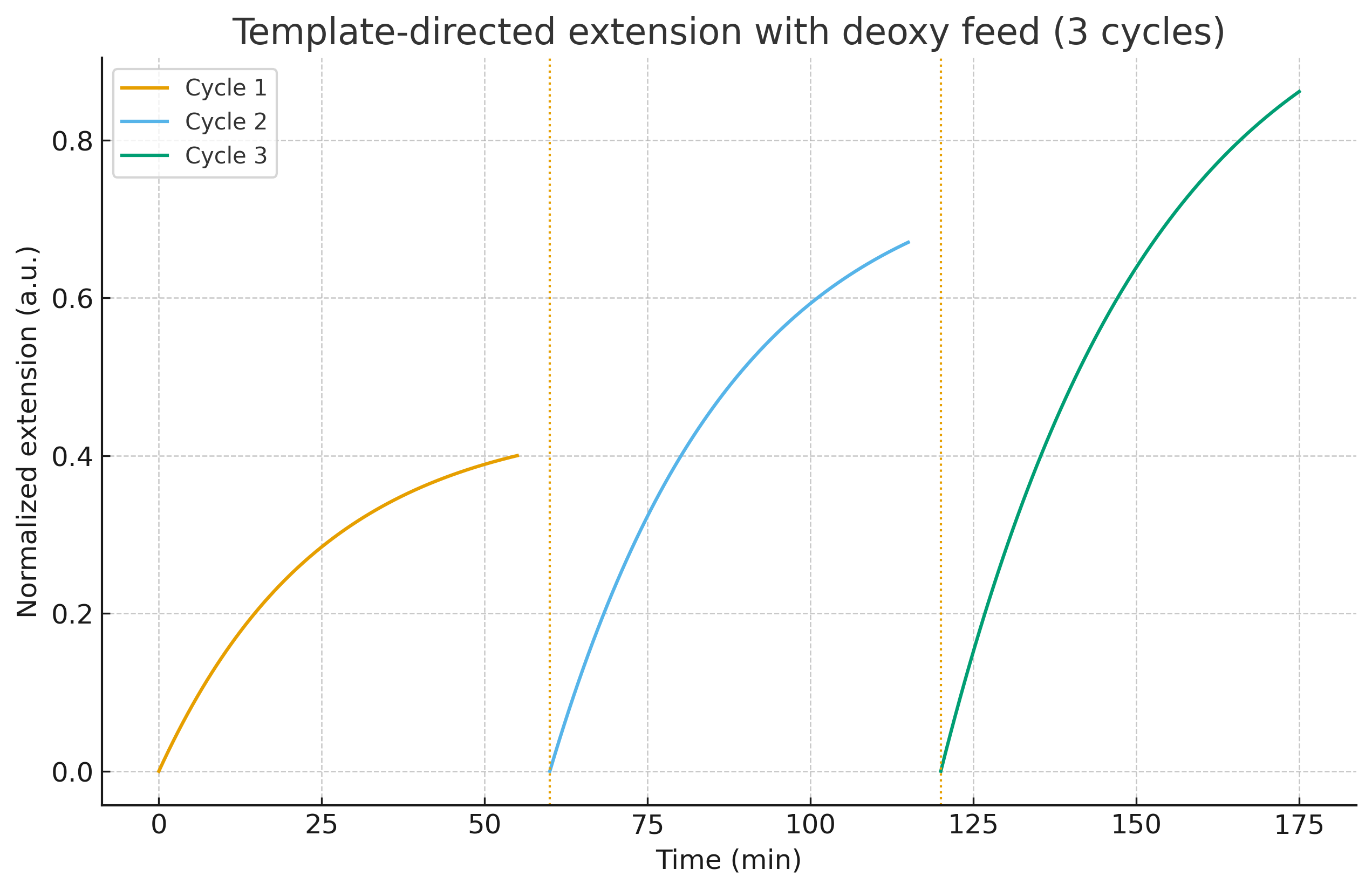
B) Copy with variation (mixed rN+dN, cyclic regime)
-
Setup. Non-enzymatic template-directed extension with activated monomers (rN + dN mix from A), 3 wet–dry (or thermal) cycles; pH 7.5–8.5; Mg²⁺/Fe²⁺ within protocell-compatible ranges.
-
Readout. In-cycle fluorescence/gel proxy for growth; post-cycle LC-MS (or enzymatic digest + LC-MS) to compute fraction deoxy per strand.
- Cleanup before copying. Verify residual sulfite ≤ 0.5–1 mM and Fe ≤ 10 µM (ICP or colorimetric) prior to any vesicle/copying step; quench/remove as needed.
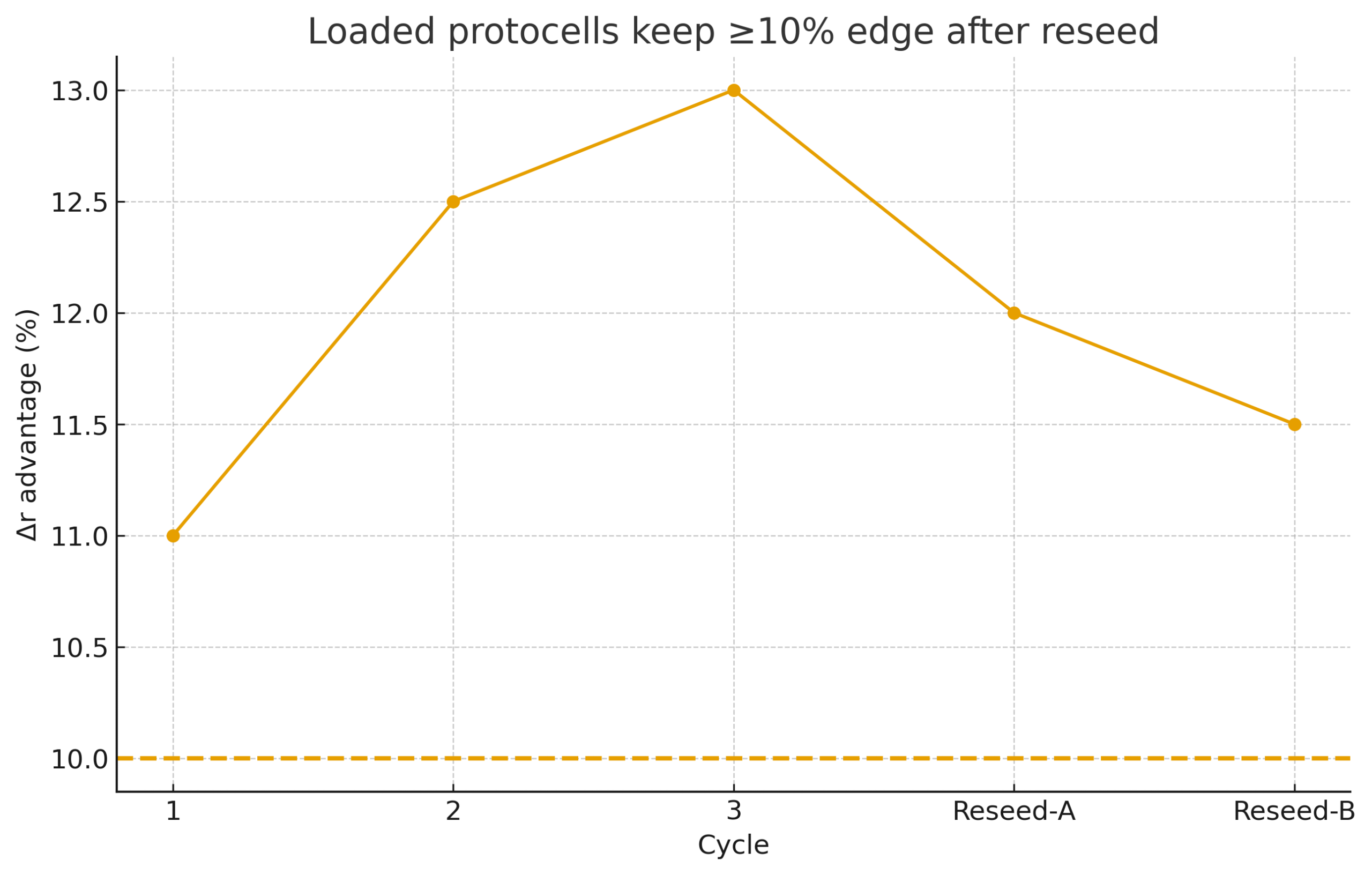
D) Selection at the compartment level
-
Setup. Compete loaded vs control protocells across cycles; re-seed twice.
-
Metric. Growth-rate advantage Δr for loaded cohort.
-
Pass if: Δr ≥ 10% and the edge persists after reseed.
Pre-registered thresholds (one-page version)
-
A (Feed): UV-B-only bandpass; dose 20–70 J cm⁻²; dA/dI ≥5–10 µM; dN:rN ≥0.3.
-
B (Copying): 3 cycles with measurable extension; no cycle stalls.
-
C (Trend): Deoxy-fraction slope >0 (95% CI).
-
D (Selection): Δr ≥10% persists after ≥2 reseeds.
- E (Cleanup): residual sulfite ≤ 0.5–1 mM; Fe ≤ 10 µM before copying/vesicles.
What would falsify the “deoxy takeover” step?
-
The UV-B/sulfite feed never clears the dN gate.
-
Copying runs, but deoxy fraction does not rise (slope ≤0).
-
Loaded protocells lose the edge after reseed.
Any one of these fails means we tighten or drop the step; no miracle hand-waves.
How this fits the program
-
If Part 2 (Interface-Heartbeat) passes, we’ve shown the wall can power autocatalysis.
-
If Part 3 (Protolife) passes, we’ve shown copying + selection in compartments.
-
Then Part 4 asks whether daylight chemistry can steer that system toward DNA-like building blocks—again, with pre-registered gates.